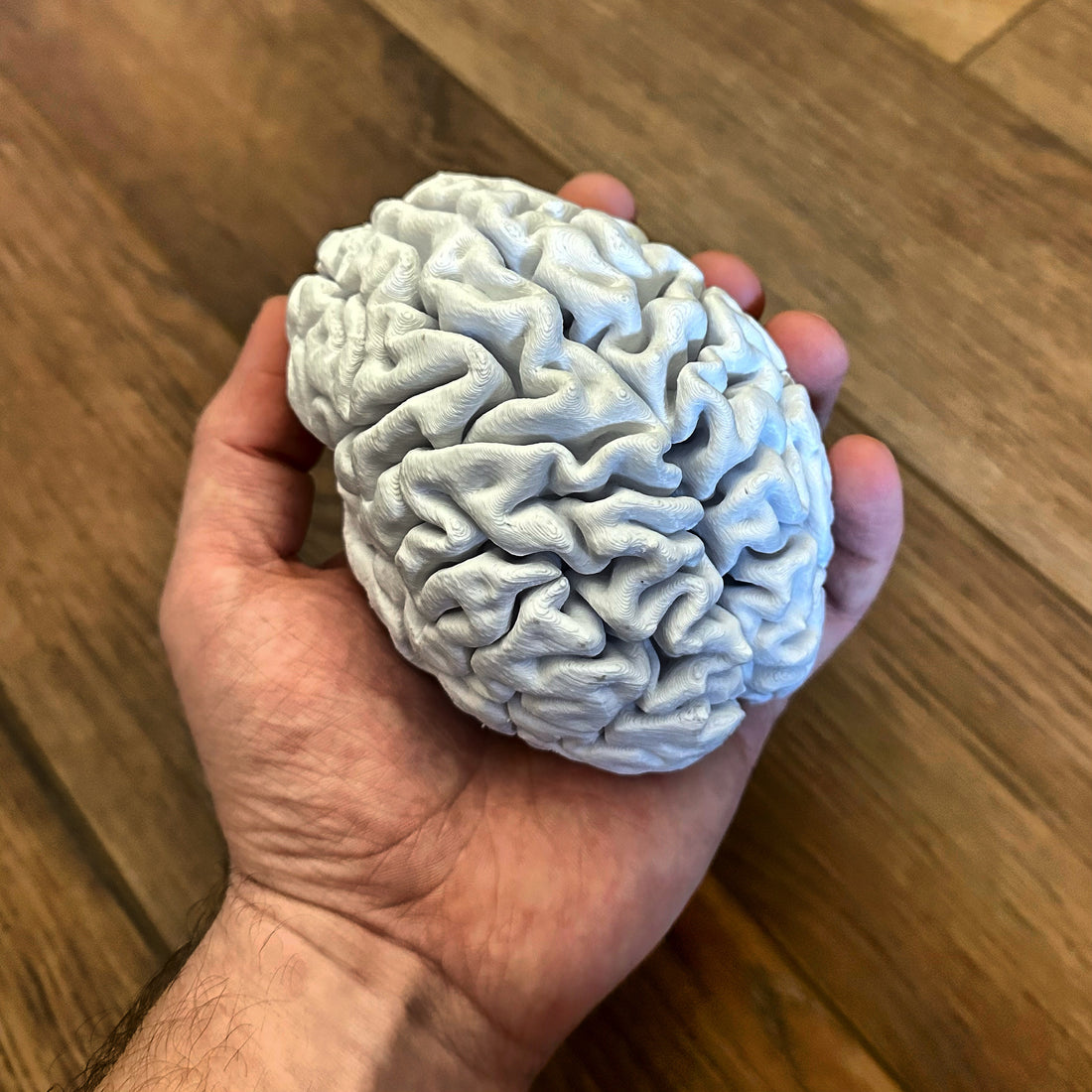
3D Printing Your Brain
Share

It's weird, isn't it? With a little bit of time and some PLA, you can hold your own brain in the palm of your hand. Everything that makes you, you - and it's right there.
Turns out, it's not that difficult to 3D print your own brain. The hardest part is getting the MRI or CT scan, and usually you don't necessarily want a reason to have one of those. Nonetheless, if you happen to have had one in the last couple years, odds are you can get a copy of those files for your own use. Some hospitals or clinics charge a small fee for a CD with your records on it, but it's a small price to pay considering what the MRI itself costs!
I found a few guides scattered across the internet, but most were outdated, used defunct software, or left out a lot of crucial details which made this a lengthy process for me at first. I've gone ahead and made this guide to be more detailed so hopefully anybody can achieve the same results I did. Feel free to comment below with any questions, and hopefully if I can't answer them, somebody else can.
Prerequisites
So here's what you'll need in addition to a copy of your scans. For reference, I did this on an M1 MacBook Pro, so this guide will focus on that specific operating system, but it should be similar on Linux or Windows.
- Slicer - a free and open-source platform for analyzing and understanding medical image data
- FreeSurfer - an open source neuroimaging toolkit for processing, analyzing, and visualizing human brain MR images
- MeshLab - an open source system for processing and editing 3D triangular meshes
Here's the Plan
The process can be summarized in four parts. First, we'll use Slicer to review the scans and pick the best one to export. Then, after exporting the scan in Slicer, we will use FreeSurfer to generate a printable 3D model out of the MRI slices. This process takes a very long time. For reference, it took about ten hours on my M1 MacBook Pro. After the model is generated, we'll clean it up and smooth it out (don't worry, not that smooth) for better printing using MeshLab. Once that's finished, the STL file will be ready for printing!
So if you're ready to proceed, just install the programs listed above and follow along!
Step 1: Gather Your Images
First thing's first - you'll have to get your scans/images together and see what you've got. Most hospitals or imaging clinics will give you a CD with your scans on it. If this is the case, I would suggest copying the files to a hard drive, locally if you have the room or a portable hard drive otherwise. This will be much faster to work with than a CD.
While the most common format for MRI scans is DICOM (.dcm), some organizations might export your scans in a different format. This guide assumes your scans are in DICOM format, but they'll be converted to NifTI (.nii) format later. If your scans are already in NifTI format, compressed (.nii.gz) or uncompressed (.nii) then you're already a step ahead! My scan happened to be in DICOM format:
Step 2: Examine Your Scans in Slicer
Once you have your image files copied to a hard drive, open Slicer. If your images were in NiFTI format, click the Add Data  button, then choose either the Choose Directory to Add or Choose File(s) to Add button depending on how many files you have. Browse to your file(s) or directory, select them, and click the Open button, followed by the OK button to finish importing them.
button, then choose either the Choose Directory to Add or Choose File(s) to Add button depending on how many files you have. Browse to your file(s) or directory, select them, and click the Open button, followed by the OK button to finish importing them.
If your files were DICOM like mine were, then you're going to click the Add DICOM Data  button. Browse to the folder with your image files, and click the Import button. You should see a notification at the bottom of the window that your information was imported:
button. Browse to the folder with your image files, and click the Import button. You should see a notification at the bottom of the window that your information was imported:
From here, you should see your name in the Patient name pane. Click your name, then click the scan in the Study pane, and then click the Load button at the bottom of the window.
Step 3: Export a Scan from Slicer
Once your scans are loaded, you should see a preview on the right side of the Slicer window. If you have multiple scans, you can click the view button  next to each one to see which looks best. I am not a radiologist or a doctor so I'm not really sure what the difference was between all my scans, but I chose the one which looked best and most clear from all three axis.
next to each one to see which looks best. I am not a radiologist or a doctor so I'm not really sure what the difference was between all my scans, but I chose the one which looked best and most clear from all three axis.
Once you've chosen which scan you like most, right-click the scan in the PANE and click the Export to file... option. From there, change the Export format drop-down to NifTI (.nii.gz) format and make sure the Compress checkbox is checked/ticked! Click the ... button in the Directory: field and browse to a folder on your local hard drive to export the scans to, and click Open. Make a note of the filename, and change it if you'd like. Ensure the filename has no spaces in it so there are no issues with running commands in Terminal later on.
Finally, click the Export button.
Step 4: Process the Scan with FreeSurfer
Now that you've got a compressed NifTI file, you're ready to process the images with FreeSurfer to generate a 3D model our of the scans. You should have already installed FreeSurfer using the link above. After it's installed, though, we need to export the necessary environment paths so your computer knows where the program binaries are located so they can be run without absolute references each time.
First, open Terminal. Then, confirm the location of your FreeSurfer install. For me, it was "/Applications/freesurfer/7.3.2" at the time of writing this.
Next, we need to define the location $FREESURFER_HOME as this path. Substitute the path of your FreeSurfer installation as necessary, and use the following command:
export FREESURFER_HOME=/Applications/freesurfer/7.3.2
FreeSurfer should include a script to set the rest of the environment variables and paths. Run the script like this:
source $FREESURFER_HOME/SetUpFreeSurfer.sh
From there, we're ready to start processing. Pick a name for your "subject" which will create a child directory for this project inside the FreeSurfer directory. I picked the very creative name "mybrain" but you can substitute yours in the command below. The next command will take a very, very long time to run. As previously mentioned, mine took about ten hours on an M1 MacBook Pro. If you're using a laptop, I would suggest leaving your computer plugged in and (if using a MacBook) using an app like Amphetamine to keep it from sleeping.
Let's get this model processed by running the following command, using the path to the file you exported in Slicer:
recon-all -s mybrain -all -i /path/to/your/exported/FILE.nii.gz
If you're using macOS, you can drag-and-drop the file into the terminal to have the path pasted automatically.
Now sit back, relax, and waaaiiittttt....
Step 5: Convert the Model to STL with FreeSurfer
Once the process is finally complete, we can convert it to an STL. FreeSurfer will generate two files, one for each hemisphere of your brain, so we'll have to convert two files. Substitute your FreeSurfer path and subject name as necessary in the commands below:
cd /Applications/freesurfer/7.3.2/subjects/mybrain/surf
mris_convert rh.pial rh.pial.stl
mris_convert lh.pial lh.pial.stl
Step 6: Clean Up the STL with MeshLab
Now that we have our STL files, let's get them polished up! Open MeshLab, click Import Mesh, and browse to your subject directory where your STL files were converted above. Choose one of the STL files, use the default options if prompted, and then repeat the process to import the second STL.
You should now see a nice 3D image of your brain! Pretty cool, huh? Let's clean it up a bit for printing!
Click on Filters > Mesh Layer > Flatten Visible Layers. Click the Apply button, and check the status pane in the bottom-right corner of the window. Once you see "Merged all the layers to single mesh..." you can click the Close button.
Next, click Filters > Remeshing, Simplification, Reconstruction > Simplification: Quadratic Edge Collapse Decimation. In the Target number of faces field, I entered 200000 and was happy with the result. Click the Apply button. Again, keep an eye on the status pane until you see "Applied filter Simplification: Quadratic Edge Collapse Decimation ..." and then click the Close button.
Lastly, click Filters > Smoothing, Fairing and Deformation > Laplacian Smooth and click Apply. Once you see "Applied filter Laplacian Smooth..." in the status window, click Close.
Finally, it's time to save the finished STL for printing! Click File > Export Mesh As... and choose a folder and filename to save the finished STL. Change the file type dropdown to STL File Format and click Save.
Step 7: Print!
Load up the final STL file in your favorite slicer, enable supports, and print away!
Feel free to post some pictures of your final print in the comments below if you'd like!

1 comment
Instructions worked perfectly. I printed this on my Bambu X1C with Bambu Matte Sakura Pink filament. I’m sure it would have looked nicer if I used a smaller layer height than 0.2 but I didn’t want to wait that long for it to print and this gave the general effect.
https://1drv.ms/u/s!At3xED2bYYcBiupE_KRlX7fq681WPQ?e=8M6CGB